Comparison of two deafness gene detection methods in genetic hearing loss gene screening
-
摘要:
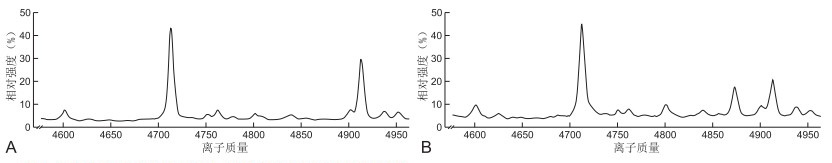
目的 探讨PCR+导流杂交法和基质辅助激光解吸电离飞行时间质谱法在遗传性耳聋基因筛查中的临床应用价值。 方法 选取杭州市第一人民医院自2016年7月—2017年7月经耳鼻咽喉科检测为听力障碍患者236例, 应用PCR+导流杂交法和基质辅助激光解吸电离飞行时间质谱法检测常见的4个耳聋相关基因: GJB2、GJB3、SLC26A4和线粒体12SrRNA的20个位点突变情况。所有样本均采用Sanger测序法进行验证。 结果 PCR+导流杂交法检测出耳聋基因突变48例, 其中GJB2基因突变率为11.44%, 杂合突变17例, 纯合突变10例; GJB3基因突变率为0.42%, 杂合突变1例; SLC26A4基因突变率为5.08%, 杂合突变10例, 纯合突变2例; 12 s RNA基因突变率为0.85%, 异质突变2例; 双杂合突变6例。基质辅助激光解吸电离飞行时间质谱法检测出耳聋基因突变53例, 其中GJB2基因突变率为11.86%, 杂合突变18例, 纯合突变10例; GJB3基因突变率为0.85%, 杂合突变2例; SLC26A4基因突变率为5.51%, 杂合突变10例, 纯合突变3例; 12 s RNA基因突变率为0.85%, 异质突变2例; 双杂合突变8例。2种方法的耳聋基因检出率分别为20.3%和22.5%, 检测结果差异无统计学意义(P>0.05)。 结论 采用合理的检测方法进行耳聋基因筛查, 是预防和降低遗传性耳聋发生的重要手段之一。 -
关键词:
- PCR+导流杂交法 /
- 基质辅助激光解吸电离飞行时间质谱法 /
- 耳聋基因 /
- 遗传性耳聋
Abstract:Objective To explore the clinical application value of PCR + flow-through hybridisation method and matrix-assisted laser desorption ionisation time-of-flight mass spectrometry in genetic deafness screening. Methods A total of 236 patients with hearing impairment were selected from the Department of Otolaryngology in Hangzhou First People's Hospital from July 2016 to July 2017. PCR + flow-guided hybridisation and matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry were used to detect 20 mutations in four common deafness-related genes: GJB2, GJB3, SLC26A4 and mitochondrial 12SrRNA. All samples were verified by Sanger sequencing. Results PCR + diversion hybridisation method detected 48 cases of deafness mutations, of which the GJB2 gene mutation rate was 11.44%, 17 heterozygous mutations and 10 homozygous mutations; the GJB3 gene mutation rate was 0.42%, 1 heterozygous mutation; the SLC26A4 mutation rate was 5.08%, 10 heterozygous mutations and 2 homozygous mutations; the 12 s RNA mutation rate was 0.85%, 2 heterogeneous mutations and 6 double heterozygous mutations. Matrix-assisted laser desorption ionisation time-of-flight mass spectrometry detected 53 cases of deafness gene mutations, of which the GJB2 gene mutation rate was 11.86%, 18 heterozygous mutations and 10 homozygous mutations; the GJB3 gene mutation rate was 0.85%, 2 heterozygous mutations; the SLC26A4 gene mutation rate was 5.51%, 10 heterozygous mutations and 3 homozygous mutations; the 12 s RNA gene mutation rate was 0.85%, 2 heterogeneous mutations and 8 double heterozygous mutations. The detection rates of deafness genes of the two methods were 20.3% and 22.5%, respectively, and no statistically significant difference was found in the detection results (P>0.05). Conclusion The use of reasonable detection methods for deafness gene screening is important to prevent and reduce the occurrence of hereditary deafness. -
表 1 耳聋基因各基因型分析结果(例)
基因 突变位点 PCR导流杂交法 基质辅助激光解吸电离飞行时间质谱法 杂合/异质 纯合/同质 杂合/异质 纯合/同质 GJB2 235delC 16 10 16 10 176 del16 0 0 0 0 299delAT 1 0 2 0 35delG 0 0 0 0 SLC26A4 IVS7-2A>G 8 2 6 3 2168 A>G 2 0 2 0 1229 C>T - - 1 0 IVS15+5G - - 1 0 12S rRNA mt1494 C>T 0 0 0 0 mt1555 A>G 2 0 2 0 GJB3 538 C>T 1 0 1 0 547 G>A - - 1 0 双杂合型 GJB2 235delC+GJB2 176del16 2 0 2 0 GJB2 235delC+GJB2299delAT 3 0 3 0 SLC26A4IVS7-2A>G+SLC26A42168A>G 1 0 1 0 SLC26A4IVS7-2A>G+SLC26A4IVS15+5G - - 1 0 GJB2 235delC+SLC26A4-1229C>T - - 1 0 注:“-”表示PCR+导流杂交法试剂盒未包含此基因突变位点。 表 2 2种耳聋基因检测方法的耳聋基因检出率(例)
方法 突变 未突变 检出率(%) PCR+导流杂交法 48 188 20.3 基质辅助激光解吸电离飞行时间质谱法 53 183 22.5 注:2种检测方法比较,χ2=0.289,P=0.591。 -
[1] 李淑娟, 刘晓雯, 陈兴健, 等. 常见综合征型耳聋临床表型及相关基因研究进展[J]. 中华耳科学杂志, 2018, 16(3): 375-381. doi: 10.3969/j.issn.1672-2922.2018.03.023 [2] 柴福, 马世博, 沈珺. 非综合征型聋患儿及其家庭成员常见耳聋基因变异分析[J]. 中国耳鼻咽喉颅底外科杂志, 2018, 24(5): 459-464. https://www.cnki.com.cn/Article/CJFDTOTAL-ZEBY201805019.htm [3] 王屹, 陈蕾, 刘志忠, 等. 318例中国汉族非综合征性耳聋患者基因突变谱分析[J]. 中国康复理论与实践, 2016, 22(12): 1451-1454. doi: 10.3969/j.issn.1006-9771.2016.12.019 [4] 刘闽, 胥亮, 刘水霞, 等. 广西地区222例感音神经性聋患者常见耳聋基因筛查结果分析[J]. 听力学及言语疾病杂志, 2017, 25(1): 5-8. doi: 10.3969/j.issn.1006-7299.2017.01.002 [5] 刘双双, 牛玉萍, 孙越, 等. 山东省非综合征型耳聋患者十四项遗传性耳聋基因突变筛查结果分析[J]. 山东大学耳鼻喉眼学报, 2016, 30(4): 6368. https://www.cnki.com.cn/Article/CJFDTOTAL-SDYU201604015.htm [6] 查树伟, 查佶, 傅雅丽, 等. 耳聋基因检测技术应用和研究现状[J]. 中国生育健康杂志, 2018, 29(5): 494-497. doi: 10.3969/j.issn.1671-878X.2018.05.027 [7] DAI Z Y, SUN B C, HUANG S S, et al. Correlation analysis of phenotype and genotype of GJB2 in patients with non-syndromic hearing loss in China[J]. Gene, 2015, 570(2): 272-276. doi: 10.1016/j.gene.2015.06.038 [8] 张昊昱, 张宁, 张华, 朱俊真. 耳聋基因检测在遗传性耳聋诊断及遗传咨询中的应用[J]. 中华耳科学杂志, 2016, 14(5): 639-643. doi: 10.3969/j.issn.1672-2922.2016.05.017 [9] 林文津, 郭舜民, 徐小妹, 等. 福建省606例非综合征型聋患者常见聋病基因突变检测结果分析[J]. 听力学及言语疾病杂志, 2018, 26(4): 371-374. doi: 10.3969/j.issn.1006-7299.2018.04.009 [10] 张彦, 刘晶晶, 叶秋萍, 等. 聚合酶链反应-反向点杂交法检测遗传性耳聋相关基因的热点突变[J]. 中华妇幼临床医学杂志(电子版), 2018, 14(3): 270-276. doi: 10.3877/cma.j.issn.1673-5250.2018.03.004 [11] 戴林桐, 卿丽华, 孙丹洋, 等. 基因芯片法检测72例非综合征性耳聋患者基因突变的研究[J]. 四川医学, 2016, 37(6): 602-604. https://www.cnki.com.cn/Article/CJFDTOTAL-SCYX201606010.htm [12] 韩跃峰, 许彬彬, 刘学宝. 江淮地区128例感音神经性聋患者常见耳聋基因检测结果分析[J]. 中华全科医学, 2019, 17(6): 906-908, 1013. https://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201906006.htm [13] 王芳, 陈小婉, 徐百成, 等. 2598例非综合征型耳聋患者基因型与临床表型的分析[J]. 中华耳科学杂志, 2016, 14(6): 747-752. doi: 10.3969/j.issn.1672-2922.2016.06.010 [14] 李俎怡, 何蓉. 339例非综合征型耳聋患者致病基因突变分析[J]. 中华耳科学杂志, 2019, 17(6): 910-915. doi: 10.3969/j.issn.1672-2922.2019.06.021 [15] 窦晓宁, 徐荣华, 高玲, 等. 潍坊地区非综合征型耳聋患者遗传性耳聋基因突变情况分析[J]. 山东医药, 2019, 59(29): 49-52. doi: 10.3969/j.issn.1002-266X.2019.29.014 [16] 于晓宇, 林妘, 许军, 等. 135例大前庭导水管耳聋患者SLC26A4基因突变分析[J]. 中华耳科学杂志, 2018, 16(2): 160-164. doi: 10.3969/j.issn.1672-2922.2018.02.007 [17] 余红, 杨晶群, 吴长划, 等. 绍兴地区131名耳聋患儿常见致聋基因检测结果分析[J]. 中华全科医学, 2018, 16(10): 1683-1685. https://www.cnki.com.cn/Article/CJFDTOTAL-SYQY201810029.htm [18] 唐燕青, 文春秀, 何升, 等. 21 386例新生儿听力筛查与聋病易感基因联合筛查结果分析[J]. 中国优生与遗传杂志, 2017, 25(10): 66-67, 102. https://www.cnki.com.cn/Article/CJFDTOTAL-ZYYA201710027.htm [19] 孙毅, 孙丽丽, 潘持国, 等. 918例听障人群耳聋基因筛查结果分析[J]. 中国听力语言康复科学杂志, 2019, 17(6): 426-429. https://www.cnki.com.cn/Article/CJFDTOTAL-TLKF201906011.htm [20] LUO S, VALENCIA C A, ZHANG J, et al. Biparental inheritance of mitochondrial dna in humans[J]. Proc Natl Acad Sci USA, 2018, 115(51): 13039-13044. doi: 10.1073/pnas.1810946115 -





 下载:
下载: