Expression and significance of SEC61G in invasive breast cancer
-
摘要:
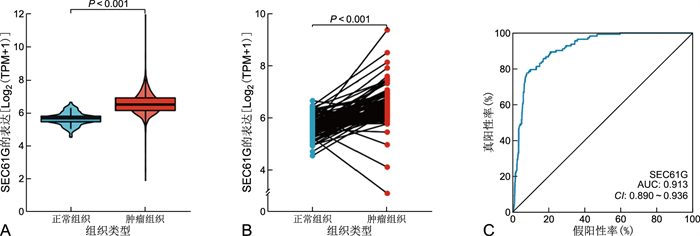
目的 利用癌症基因组图谱(TCGA)数据库中的数据分析SEC61G在浸润性乳腺癌中的表达情况及其与临床预后的关系。 方法 从TCGA数据库中收集1 222个浸润性乳腺癌患者的RNAseq数据,用Wilcoxon秩和检验分析SEC61G在正常组织和肿瘤组织中表达的差异,采用logistic回归分析SEC61G的表达与临床病理特征的关系,使用Kaplan-Meier方法和Cox回归分析评估SEC61G在预后中的作用。 结果 浸润性乳腺癌组织中SEC61G的表达明显高于正常乳腺组织(P < 0.001)。T分期(P=0.028)、N分期(P=0.009)、病理分期(P=0.003)、ER状态(P < 0.001)、PR状态(P < 0.001)、HER2状态(P=0.025)、组织学类型(P < 0.001)与SEC61G表达显著相关。单因素Cox回归分析显示SEC61G高表达与DSS有相关性(P < 0.001,HR=0.426,95% CI=0.272~0.668)。多因素Cox分析显示SEC61G高表达是疾病特异生存率(DSS)的独立危险因素(P=0.009,HR=0.479,95% CI=0.276~0.831)。SEC61G的表达与浸润性乳腺癌的免疫浸润程度有关(P < 0.05)。各组的K-M曲线显示肿瘤组织中SEC61G表达越高浸润性乳腺癌患者的预后越差(P < 0.05)。 结论 SEC61G高表达与浸润性乳腺癌预后不良有关,可以作为预测浸润性乳腺癌患者生存有效的生物标志物。 Abstract:Objective To analyze the expression of SEC61G in invasive breast cancer from tumor Genome Map (TCGA) database and its relationship with clinical prognosis. Methods The RNAseq data of 1 222 patients with invasive breast cancer were collected from TCGA database. Wilcoxon rank sum test was used to analyze the difference of SEC61G expression between normal and tumor tissues. Logistic regression was used to analyze the relationship between SEC61G expression and clinicopathological features. Kaplan-Meier method and Cox regression analysis were used to evaluate the role of SEC61G in prognosis. Results The expression of SEC61G in invasive breast cancer was significantly higher than that in normal breast tissue (P < 0.001). T stage (P=0.028), N stage (P=0.009), pathological stage (P=0.003), ER status (P < 0.001), PR status (P < 0.001), HER2 status (P=0.025) and histological type (P < 0.001) were significantly correlated with SEC61G expression. Univariate Cox regression analysis showed that the high expression of SEC61G was associated with DSS (P < 0.001, HR=0.426, 95% CI: 0.272-0.668). Multivariate Cox analysis showed that high expression of SEC61G was an independent risk factor for DSS (P=0.009, HR=0.479, 95% CI: 0.276-0.831). The K-M curve of each group showed that the higher the expression of SEC61G in tumor tissue, the worse the prognosis of invasive breast cancer patients (P < 0.05). Conclusion The high expression of SEC61G is related to the poor prognosis of invasive breast cancer and can be used as an effective biomarker to predict the survival of patients with invasive breast cancer. -
Key words:
- SEC61G /
- Invasive breast cancer /
- Prognosis
-
表 1 TCGA数据库中SEC61G高表达组和低表达组的基线特征[例(%)]
Table 1. Baseline characteristics of SEC61G high expression group and low expression group in TCGA database [cases (%)]
临床特征 分类 SEC61G高表达 SEC61G低表达 χ2值 P值 总数 541 542 T分期 T1 154(28.6) 123(22.7) 11.862 0.008 T2 297(55.1) 332(61.4) T3 77(14.3) 62(11.5) T4 11(2.0) 24(4.4) N分期 N0 266(50.3) 248(46.4) 8.491 0.037 N1 184(34.8) 174(32.5) N2 52(9.8) 64(12.0) N3 27(5.1) 49(9.2) M分期 M0 462(98.7) 440(96.9) 2.729 0.099 M1 6(1.3) 14(3.1) 病理分期 Ⅰ期 108(20.6) 73(13.6) 14.313 0.003 Ⅱ期 301(57.4) 318(59.3) Ⅲ期 111(21.2) 131(24.4) Ⅳ期 4(0.8) 14(2.6) 组织学类型 浸润性导管癌 353(72.3) 419(85.7) 25.452 < 0.001 浸润性小叶癌 135(27.7) 70(14.3) PR状态 阴性 132(25.2) 210(41.1) < 0.001a 未确定 1(0.2) 3(0.6) 阳性 390(74.6) 298(58.3) ER状态 阴性 79(15.1) 161(31.4) < 0.001a 未确定 2(0.4) 0 阳性 442(84.5) 351(68.6) HER2状态 阴性 295(80.4) 263(73.1) 5.471 0.065 未确定 5(1.4) 7(1.9) 阳性 67(18.3) 90(25.0) 注:TCGA数据库中不是所有的RNAseq都有临床信息,存在1个患者测多次的情况。TCGA数据库中不是所有的临床变量的信息都完全,或多或少存在有缺失。a为采用Fisher精确检验。 表 2 SEC61G的表达与临床病理特征的关系
Table 2. Relationship between expression of SEC61G and clinicopathological features
临床特征 数目 OR(95% CI) P值 T分期(T2~4 vs. T1) 1 080 1.359(1.034~1.791) 0.028 N分期(N0~1 vs. N2~3) 1 064 0.656(0.477~0.898) 0.009 M分期(M1 vs. M0) 922 2.450(0.974~6.970) 0.069 病理分期(Ⅰ vs. Ⅱ~Ⅳ) 1 060 0.607(0.437~0.839) 0.003 年龄(>60岁vs. ≤60岁) 1 083 0.967(0.761~1.229) 0.786 PR状态(阳性vs.阴性) 1 030 0.480(0.368~0.625) < 0.001 ER状态(阳性vs.阴性) 1 033 0.390(0.287~0.526) < 0.001 HER2状态(阳性vs.阴性) 715 1.507(1.056~2.159) 0.025 原发肿瘤部位(右vs.左) 1 083 0.859(0.677~1.091) 0.213 组织学类型(浸润性小叶癌vs.浸润性导管癌) 977 0.437(0.315~0.601) < 0.001 表 3 DSS的单因素Cox回归分析
Table 3. Univariate Cox regression analysis of DSS
临床特征 数目 HR(95% CI) P值 T分期(T2~4 vs.T1) 1 059 0.562(0.326~0.968) 0.038 N分期(N0~1 vs.N2~3) 1 044 2.462(1.502~4.036) < 0.001 病理分期(Ⅰ vs.Ⅱ~Ⅳ) 1 041 3.396(1.478~7.803) 0.004 PR状态(阳性vs.阴性) 1 010 1.926(1.238~2.997) 0.004 ER状态(阳性vs.阴性) 1 013 1.788(1.122~2.850) 0.015 组织类型(小叶癌vs.导管癌) 958 2.125(1.018~4.434) 0.045 SEC61G(高表达vs.低表达) 1 062 0.426(0.272~0.668) < 0.001 表 4 DSS的多因素Cox回归分析
Table 4. Multivariate Cox regression analysis of DSS
临床特征 数目 HR(95%CI) P值 T分期(T2~4 vs.T1) 1 059 1.048(0.444~2.474) 0.914 N分期(N0~1 vs.N2~3) 1 044 3.337(1.911~5.826) < 0.001 病理分期(Ⅰ vs. Ⅱ~Ⅳ) 1 041 1.847(0.574~5.939) 0.303 PR状态(阳性vs.阴性) 1 010 1.598(0.784~3.257) 0.197 ER状态(阳性vs.阴性) 1 013 1.151(0.537~2.466) 0.717 组织类型(小叶癌vs.导管癌) 958 2.067(0.858~4.976) 0.105 SEC61G(高表达vs.低表达) 1 062 0.479(0.276~0.831) 0.009 -
[1] NICHOLS H B, SCHOEMAKER M J, CAI J, et al. Breast cancer risk after recent childbirth: A pooled analysis of 15 prospective studies[J]. Ann Intern Med, 2019, 170(1): 22-30. doi: 10.7326/M18-1323 [2] PARK J H, KIM J H. Pathologic differential diagnosis of metastatic carcinoma in the liver[J]. Clin Mol Hepatol, 2019, 25(1): 12-20. doi: 10.3350/cmh.2018.0067 [3] CHEN Y T, NING J L, CAO W T, et al. Research progress of TXNIP as a tumor suppressor gene participating in the metabolic reprogramming and oxidative stress of cancer cells in various cancers[J]. Front Oncol, 2020, 10: 568574. DOI: 10.3389/fonc.2020.568574. [4] WANG H H, HSU Y H, CHANG M S. IL-20 bone diseases involvement and therapeutic target potential[J]. J Biomed Sci, 2018, 25(1): 38. doi: 10.1186/s12929-018-0439-z [5] SUN X, JOHNSON J, ST JOHN J C. Global DNA methylation synergistically regulates the nuclear and mitochondrial genomes in glioblastoma cells[J]. Nucleic Acids Res, 2018, 46(12): 5977-5995. doi: 10.1093/nar/gky339 [6] LIANG L F, HUANG Q W, GAN M, et al. High SEC61G expression predicts poor prognosis in patients with Head and Neck Squamous Cell Carcinomas[J]. J Cancer, 2021, 12(13): 3887-3899. doi: 10.7150/jca.51467 [7] LINXWEILER M, SCHICK B, ZIMMERMANN R. Let's talk about Secs: Sec61, Sec62 and Sec63 in signal transduction, oncology and personalized medicine[J]. Signal Transduct Target Ther, 2017, 2: 17002. DOI: 10.1038/sigtrans.2017.2. [8] BINDEA G, MLECNIK B, TOSOLINI M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer[J]. Immunity, 2013, 39(4): 782-795. doi: 10.1016/j.immuni.2013.10.003 [9] ZHOU J, SU C M, CHEN H A, et al. Cryptanshinone inhibits the glycolysis and inhibits cell migration through PKM2/beta-catenin axis in breast cancer[J]. Onco Targets Ther, 2020, 13: 8629-8639. doi: 10.2147/OTT.S239134 [10] MENG H, JIANG X W, WANG J, et al. SEC61G is upregulated and required for tumor progression in human kidney cancer[J]. Mol Med Rep, 2021, 23(6): 427. doi: 10.3892/mmr.2021.12066 [11] STEVEN A, SELIGER B. The role of immune escape and immune cell infiltration in breast cancer[J]. Breast Care (Basel), 2018, 13(1): 16-21. doi: 10.1159/000486585 [12] SHI Y N, LIANG Y B, WANG W, et al. SEC61G identified as a prognostic biomarker of head and neck squamous cell carcinoma[J]. Eur Arch Otorhinolaryngol, 2021, 279(4): 2039-2048. [13] MA J J, HE Z X, ZHANG H W, et al. SEC61G promotes breast cancer development and metastasis via modulating glycolysis and is transcriptionally regulated by E2F1[J]. Cell Death Dis, 2021, 12(6): 550. doi: 10.1038/s41419-021-03797-3 [14] SEOANE J A, KIRKLAND J G, CASWELL-JIN J L, et al. Chromatin regulators mediate anthracycline sensitivity in breast cancer[J]. Nat Med, 2019, 25(11): 1721-1727. doi: 10.1038/s41591-019-0638-5 [15] LI X M, LIU L, GOODALL G J, et al. A novel single-cell based method for breast cancer prognosis[J]. PLoS Comput Biol, 2020, 16(8): e1008133. DOI: 10.1371/journal.pcbi.1008133. [16] WILK G, BRAUN R. regQTLs: Single nucleotide polymorphisms that modulate microRNA regulation of gene expression in tumors[J]. PLoS Genet, 2018, 14(12): e1007837. DOI: 10.1371/journal.pgen.1007837. [17] PASHAYAN N, ANTONIOU A C, IVANUS U, et al. Personalized early detection and prevention of breast cancer: ENVISION consensus statement[J]. Nat Rev Clin Oncol, 2020, 17(11): 687-705. doi: 10.1038/s41571-020-0388-9 [18] VAN DER LEUN A M, THOMMEN D S, SCHUMACHER T N. CD8(+) T cell states in human cancer: Insights from single-cell analysis[J]. Nat Rev Cancer, 2020, 20(4): 218-232. doi: 10.1038/s41568-019-0235-4 -





 下载:
下载: